

- #CYTOSCAPE LINUX MAC OS X#
- #CYTOSCAPE LINUX INSTALL#
- #CYTOSCAPE LINUX UPDATE#
- #CYTOSCAPE LINUX SOFTWARE#
- #CYTOSCAPE LINUX LICENSE#
#CYTOSCAPE LINUX LICENSE#
Availability and implementation: The plug-in (running on Windows, Linux, or Mac OS) and its tutorial (both in written and video forms) can be downloaded freely under the terms of the MIT license from. Importantly, the tool gives a quality score of the network visualization by calculating the information loss between the input data and the visual representation showing a 3- to 25-fold improvement over conventional methods. The plug-in provides an efficient visualization of network modules, which represent major protein complexes in protein-protein interaction and signalling networks.
Upgrade to Cytoscape.js 2.7.15 or use one of the included workarounds Visualizing Glycolysis with Cytoscape.js Creating an animated flowchart for glycolysis. Results: We present the EntOptLayout Cytoscape plug-in based on a recently developed network representation theory. Take the first steps toward using RINalyzer here.Motivation: Network visualizations of complex biological datasets usually result in 'hairball' images, which do not discriminate network modules. Now PDB files can be loaded in Chimera through the RINalyzer menu options. Save the new preferences by clicking the option Make Current Cytoscape Properties Default at the bottom of the dialog. On a Linux machine this could be $HOME/chimera/bin, on a Windows machine:Ĭ:\Program Files\Chimera\bin, and on a Macintosh: /Applications/ Chimera.app/Contents/MacOS. Click OK and enter the path to the Chimera application. Click the Add button and enter the name of the property: Chimera.chimeraPath. Go to the Cytoscape Preferences Editor ( Edit → Preferences → Properties). Configure the path to the Chimera application in Cytoscape:. #CYTOSCAPE LINUX INSTALL#
Download and install the latest Chimera version 1.4 from the UCSF Chimera download page.Enable RINalyzer features requiring UCSF Chimera:.
#CYTOSCAPE LINUX MAC OS X#
Platform: Windows compatible, Mac OS X compatible, Linux compatible, Unix compatible.
Now start Cytoscape and find the RINalyzer menu options in the Plugins menu. Description: A Cytoscape plug-in that visualizes the non-redundant. These are summarized in the tables below. On a Macintosh and $HOME/Cytoscape_v2.8.0/plugins/ on a Linux machine. Cytoscape allows a wide variety of properties to be controlled. Download the file RINalyzer.jar here and copy it into the plugins folder of Cytoscape, e.g.Ĭ:\Program Files\Cytoscape_v2.8.0\plugins\ on a Windows machine /Applications/Cytoscape_v2.8.0/plugins/. Download and install the latest Cytoscape version 2.8.0 from the Cytoscape download page. #CYTOSCAPE LINUX UPDATE#
If Java 6 is not installed, just update it or download and install it from here. Ensure that Java 6 is installed using this test web page.Requirements and Installation for Cytoscape 2.x If no other path is specified in the structureViz Settings, this path is used for launching Chimera next time. Once Chimera has been started successfully, the last used path is saved in the session file.Go to Apps → structureViz → Launch Chimera.
#CYTOSCAPE LINUX SOFTWARE#
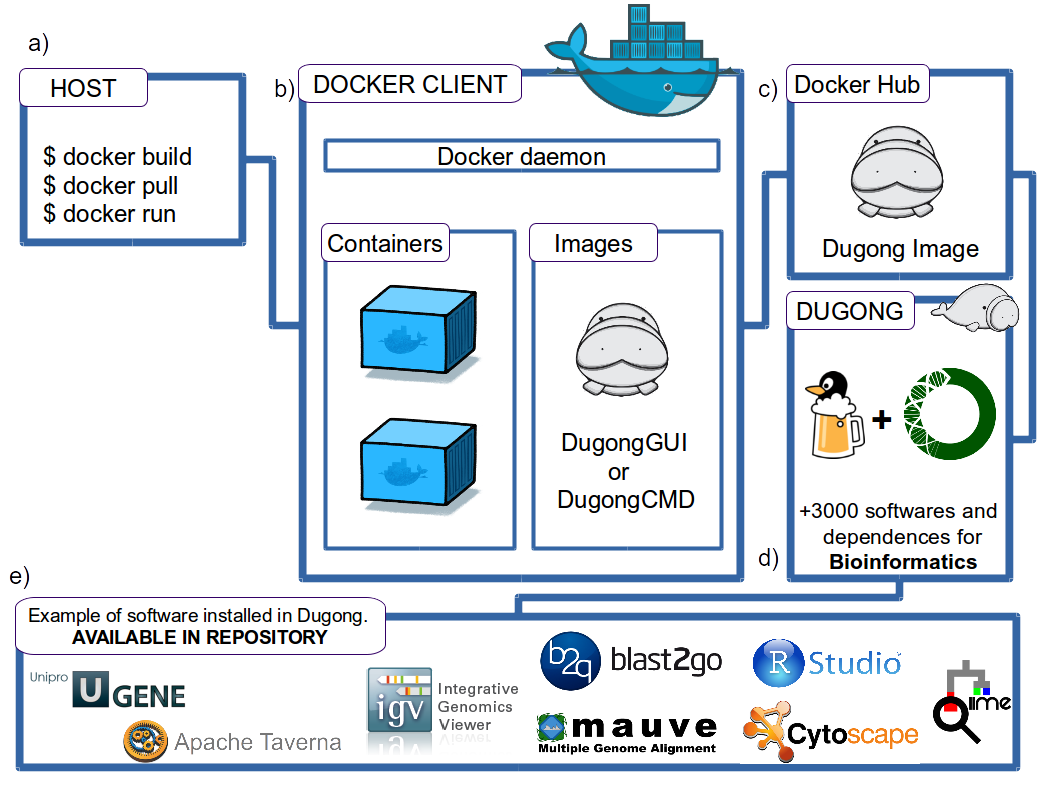
To run this software interactively in a Linux environment run the commands.
These settings are specific to each network in Cytoscape. Cytoscape is an open source bioinformatics software platform for visualizing.
If not using the default Chimera installation directory, set the path to the Chimera executable file in Apps → structureViz → Settings. Launch UCSF Chimera from within Cytoscape:. To check if the installation was successful, go to Apps to find the RINalyzer and structureViz menu options. Repeat the same action with the structureViz.jar file. button and select the RINalyzer.jar file. Open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations.  Start Cytoscape and go to the App Manager ( Apps → App Manager). Download the RINalyzer 2 app from the RINalyzer download page. Download the structureViz 2 app from the Cytoscape App Store. Install RINalyzer and structureViz app:. Thus, you must put the 'all-permissions' field in the security section or Cytoscape will crash. UCSF Chimera: Download and install the latest UCSF Chimera version (1.8 and above) from the UCSF Chimera download page. Cytoscape is currently written to require access to the local computer to read and save customization files and use system resources. Cytoscape: Download and install the latest Cytoscape version (3.1.0 and above) from the Cytoscape download page. If Java is not installed, just update it or download and install it from here. Java: Ensure that Java 6 or 7 is installed using this test web page. Requirements and Installation for Cytoscape 3.x
Start Cytoscape and go to the App Manager ( Apps → App Manager). Download the RINalyzer 2 app from the RINalyzer download page. Download the structureViz 2 app from the Cytoscape App Store. Install RINalyzer and structureViz app:. Thus, you must put the 'all-permissions' field in the security section or Cytoscape will crash. UCSF Chimera: Download and install the latest UCSF Chimera version (1.8 and above) from the UCSF Chimera download page. Cytoscape is currently written to require access to the local computer to read and save customization files and use system resources. Cytoscape: Download and install the latest Cytoscape version (3.1.0 and above) from the Cytoscape download page. If Java is not installed, just update it or download and install it from here. Java: Ensure that Java 6 or 7 is installed using this test web page. Requirements and Installation for Cytoscape 3.x







 0 kommentar(er)
0 kommentar(er)
